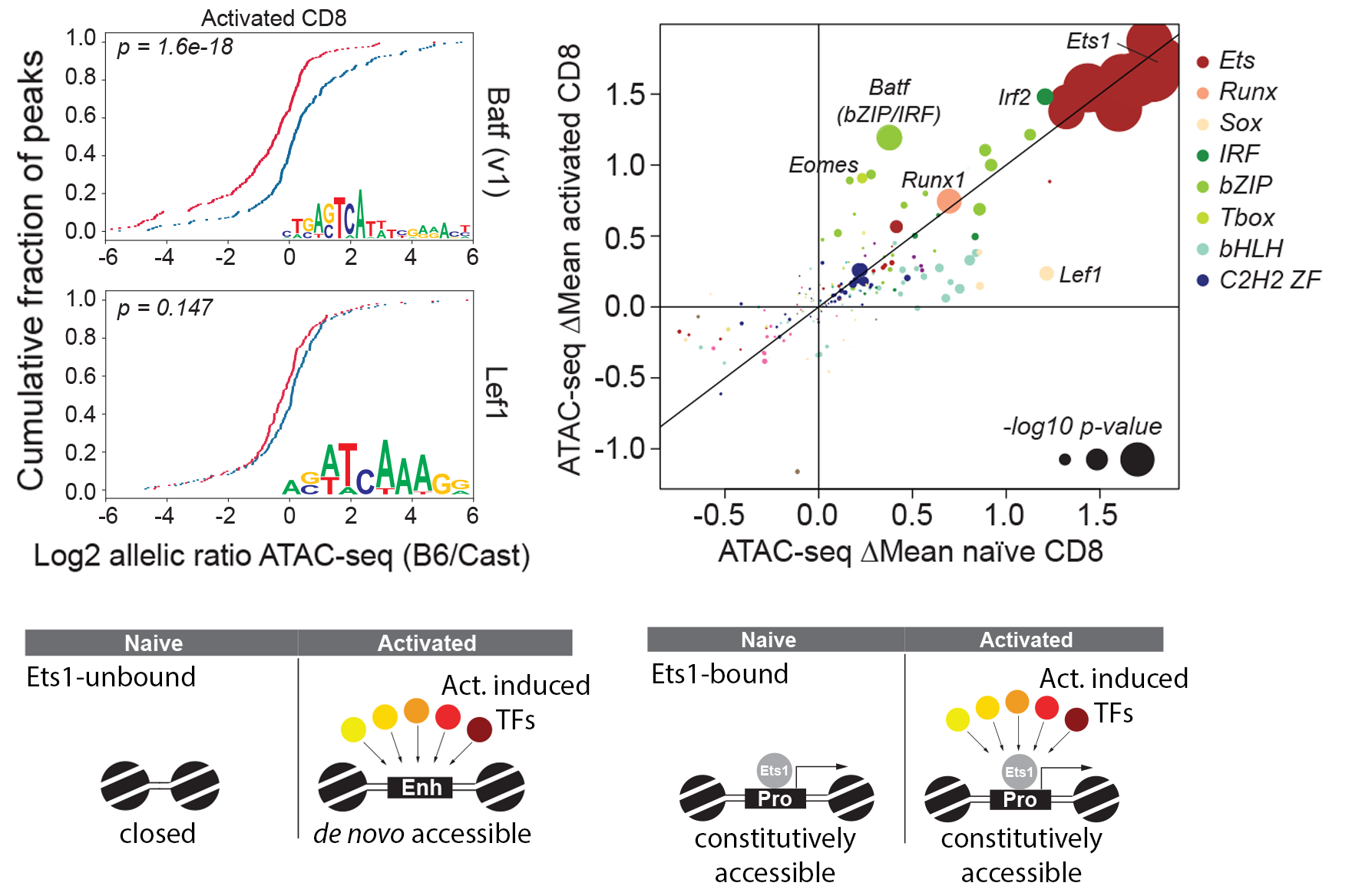
Quantitative regulation of T cell subpopulation
Our lab focuses on understanding quantitative regulation of multiple TF families governing T cell subset differentiation in response to infection. Our efforts can be split into constructing specific mouse model to leverage substantial genetic diversity of strains and establishing novel integrative strategies to dissect the joint transcriptional regulation consequences in T cell activation in a quantitative manner at bulk and single cell levels.
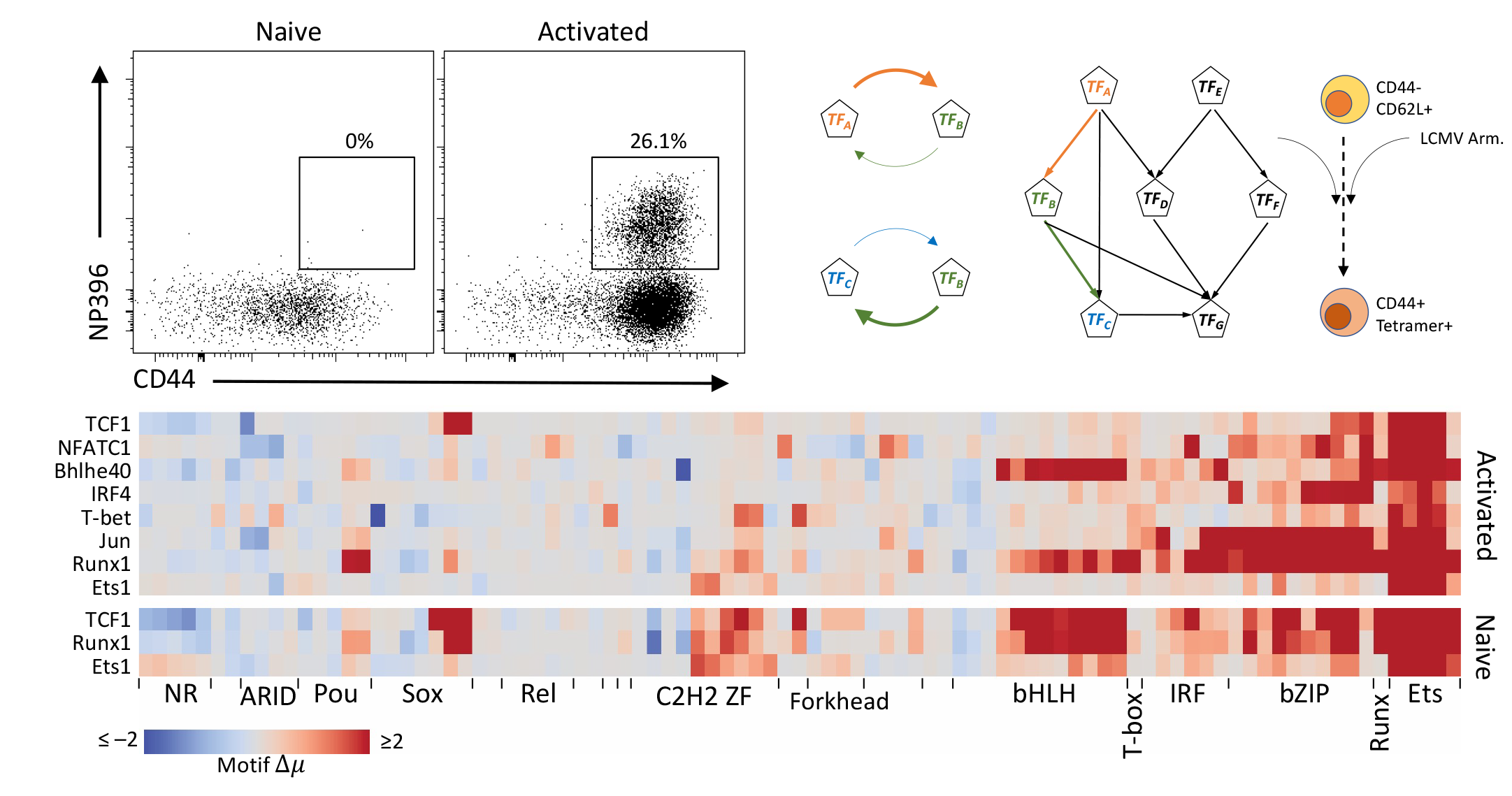
Modeling T cell differentiation
We are interested in modeling the intrinsic relation among TF binding, chromatin accessibility and gene expression in T cell development as well as T cells in tumor microenvironment. Hierarchical transcriptional network will further be built and refined with genetic perturbations in multiple T cell subtypes. The final goal is to predict the T cell fate with trained model taking TF binding redundancy and complexity into account.
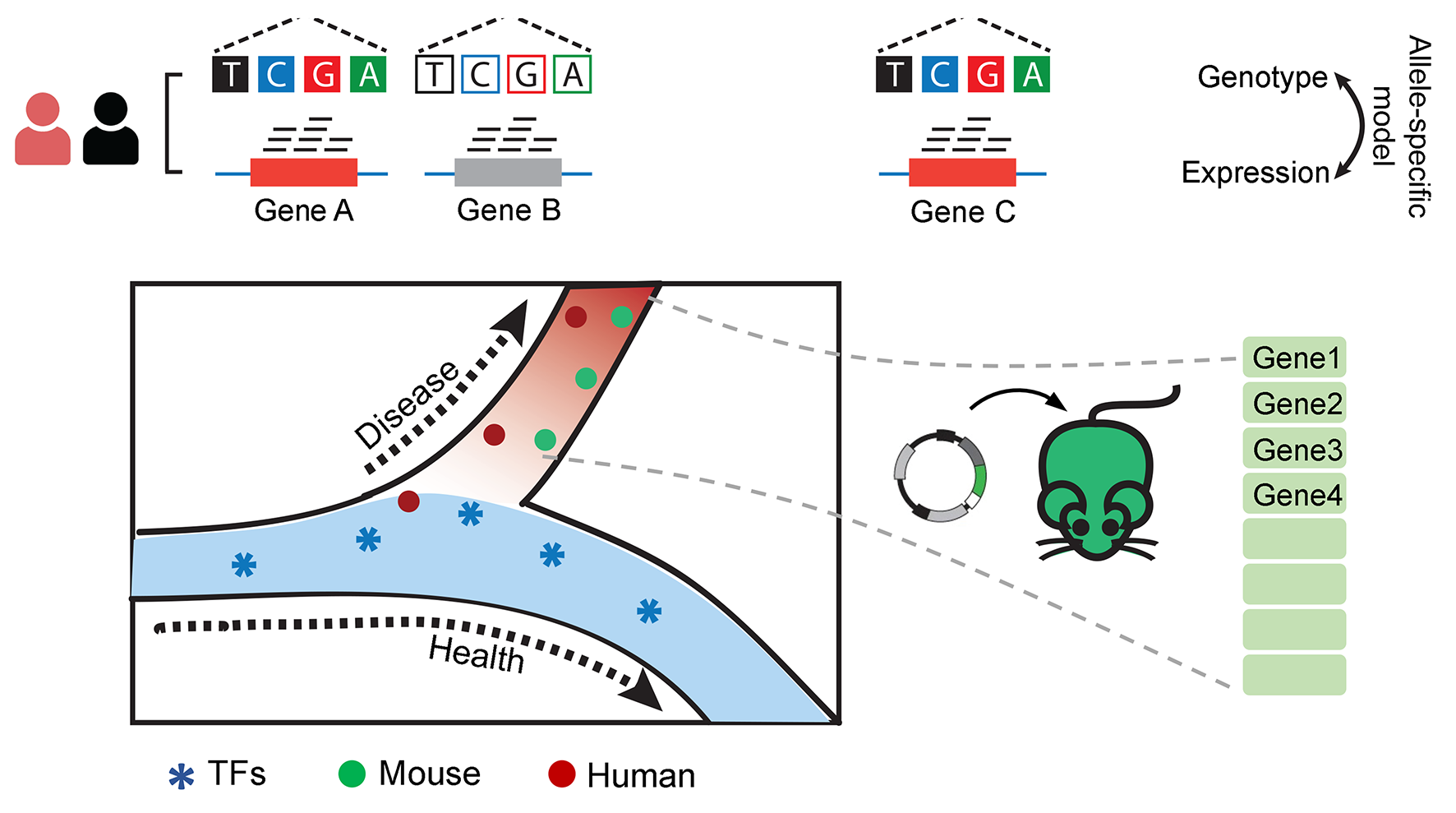
Molecular etiology of diseases of lymphatic system
We seek to reconstruct the pathological process of lymphatic disease from its normal precursor cells. To gain epigenetic and transcriptional insights, studies at three levels including healthy individuals, pre-clinical models and clinical patients will be investigated to unravel the key transcription factors and essential genes that sequentially promote or accelerate the deviation of disease status from the health developmental path.


